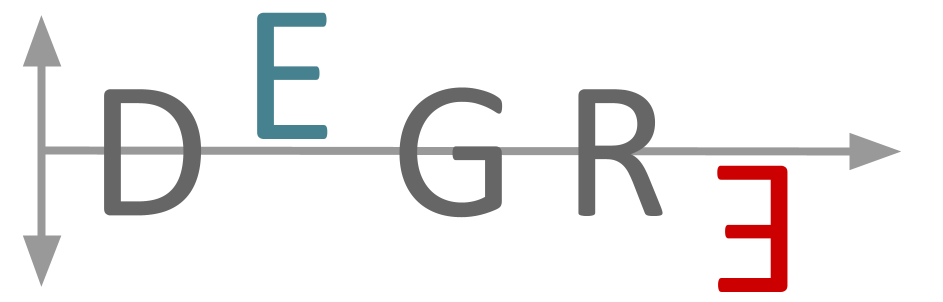
The DEGRE is an R package that aims to identify Differentially Expressed Genes (DEGs) in a pairwise manner and considers the insertion of the individuals’ random effects in the experimental design. This package has a preprocessing step responsible for filtering genes that could impair DEGs’ inference. For this purpose, DEGRE uses Generalized Linear Mixed Model (GLMM) with the negative binomial distribution.
The repository for DEGRE is at GitHub on the https://github.com/labinfo-lncc/DEGRE. In this website you can report a bug and get help.
The DEGRE package is under publication. Please cite it if you use it in your research.
You can cite it here below:
citation("DEGRE")# To cite package ‘DEGRE’ in publications use:
# Machado, et al. (2022). DEGRE: Inferring differentially expressed genes using generalized linear mixed models in the DEGRE R package. R package version 1.0.
# A BibTeX entry for LaTeX users is
# @Manual{,
# title = {Inferring differentially expressed genes using generalized linear mixed models in the DEGRE R package},
# author = {Douglas Terra Machado, Otavio Jose Bernardes Brustolini, Yasmmin Cortes Martins, Marco Antonio Grivet Mattoso Maia, Ana Tereza Ribeiro Vasconcelos},
# year = {2022},
# note = {R package version 1.0},
# }
# ATTENTION: This citation information has been auto-generated from the package
# DESCRIPTION file and may need manual editing, see ‘help("citation")’.To install the DEGRE package, you must run:
library(devtools)
install_github("labinfo-lncc/DEGRE", ref="main")The DEGRE package has several functions from the following packages:
Attention: if you are having trouble with the glmmTMB package, you can try removing and reinstalling it.
To use this package, you must have a matrix containing the read counts for each gene in each biological replicate and a design matrix containing sample information. In the count matrix, the rows represent genes, while the columns represent the biological replicates of both experimental conditions. In the design matrix, the columns represent the relevant information of the samples.
To start using the package, you need to load the library:
library("DEGRE")Then you can apply the DEGRE’s quick start function:
# Reading the count matrix and the design matrix for an example:
dir <- system.file("data", package = "DEGRE")
tab <- read.csv(file.path(dir,"count_matrix_for_example.csv"))
row.names(tab) <- tab[,1]; tab <- tab[,-1]
des <- read.csv(file.path(dir,"design_matrix_for_example.csv"))
# Running DEGRE function:
results <- DEGRE(count_matrix = tab,
p_value_adjustment = "BH",
design_matrix = des,
formula = "condition + (1|sex)")Its output (represented as the results above) is a
data.frame object. This object has the columns:
count_matrix.This function has the following arguments:
results <- DEGRE(count_matrix,
design_matrix,
formula,
p_value_adjustment)As you can see, the DEGRE function has important parameters, which are described below:
count_matrix - a data.frame object. It
receives the matrix as input.design_matrix - a data.frame object. It
receives the experimental design matrix. The sample names must be
identified in the first column. This matrix can also have more columns
with information for the fixed and the random effects for the
samples.formula - it receives fixed and random effects
descriptions.p_value_adjustment - All the P-values computed
must be corrected and the DEGRE package offers two possibilities: “BH”
(Benjamini-Hochberg) correction (default) and “BON” (Bonferroni)
correction.In a well-planned experimental design, fixed effects are features that are constant across the investigated individuals. Let’s suppose we are doing an RNA-Seq experiment to identify DEGs in whole blood between a group of people that receives a specific treatment and a group that receive only water instead of medicine. The illustration of this comparison can be seen below:

Notice that we know that all people in group A received the treatment. In contrast, all people in group B received only water. This kind of comparison in which we can control the effect between both groups comprehends the fixed effects.
Suppose that in both groups, we have some heterogeneity related to the biological sex, in which there are men and women in different proportions. The illustration of this can be seen below:

This kind of effect can be handle as a random effect to eliminate any potential bias related with variations between males and females¹²³. Other examples of random effects that can be harder to control may involve the different concentrations of biochemical molecules inside both groups implicating in the gene expression.
Thoral, Elisa, Quentin Queiros, Damien Roussel, Gilbert Dutto, Eric Gasset, David J. McKenzie, Caroline Romestaing, Jean-Marc Fromentin, Claire Saraux, and Loïc Teulier. 2021. “Changes in Foraging Mode Caused by a Decline in Prey Size Have Major Bioenergetic Consequences for a Small Pelagic Fish.” The Journal of Animal Ecology 90 (10): 2289–2301.
Kleyheeg, Erik, Bart A. Nolet, Sandra Otero-Ojea, and Merel B. Soons. 2018. “A Mechanistic Assessment of the Relationship between Gut Morphology and Endozoochorous Seed Dispersal by Waterfowl.” Ecology and Evolution 8 (22): 10857–67.
Donovan, Margaret K. R., Agnieszka D’Antonio-Chronowska, Matteo D’Antonio, and Kelly A. Frazer. 2020. “Cellular Deconvolution of GTEx Tissues Powers Discovery of Disease and Cell-Type Associated Regulatory Variants.” Nature Communications 11 (1): 955.
Here are some codes you can apply to filter specific gene IDs.
Suppose you want to filter the results related to the ENSMUSG00000000881 gene ID. Then, you can try:
filtering_gene <- results[results$ID == "ENSMUSG00000000881",]
print(filtering_gene)Applying this filtering step, you get the following result:
# ID log2FC P-value Q-value
# ENSMUSG00000000881 -3.71189 1.83797593742305e-64 7.765448e-64However, suppose you have specific gene IDs in a vector, for example:
gene_IDs_to_filter <- c("ENSMUSG00000000881",
"ENSMUSG00000002820",
"ENSMUSG00000005610",
"ENSMUSG00000015484")We can use the merge function to get the gene results. First, consider converting it into data.frame object:
gene_IDs_to_filter <- data.frame(ID = gene_IDs_to_filter)
print(gene_IDs_to_filter)# ID
# ENSMUSG00000000881
# ENSMUSG00000002820
# ENSMUSG00000005610
# ENSMUSG00000015484Then, you can use the merge function:
filtering_gene <- merge(results, gene_IDs_to_filter, by = "ID")
print(filtering_gene)# ID log2FC P-value Q-value
# ENSMUSG00000000881 -3.7118904 1.83797593742305e-64 7.765448e-64
# ENSMUSG00000002820 -4.4200312 5.10465155023083e-105 2.974780e-104
# ENSMUSG00000005610 1.4922973 2.41432633104465e-115 1.522467e-114
# ENSMUSG00000015484 -0.6455689 7.54077969856174e-06 1.098614e-05To filter the Q-value from a specific cutoff, you can do the following:
results_q_value_cutoff <- results[results$`Q-value` < 0.05,]Here we filtered based on the 5% of significance as a cutoff, but you can change it.
To check if it works properly, you can check the maximum
Q-value from results and
results_q_value_cutoff:
max(results_q_value_cutoff$`Q-value`)# 0.0493556Comparing it with the results, i.e., the data frame
without the filtering, you get:
max(results$`Q-value`)# 0.9519065In which it shows you that the filter works properly.
If you only have the gene IDs but want the gene names, you can use the biomaRt package to get ensembl information.
First, if you don’t have the biomaRt installed, please install it following the recommendations here.
Then, you must do the library:
library("biomaRt")Using the following code, you can get the gene names from the Mus musculus species:
ensembl <- useMart("ensembl",
dataset="mmusculus_gene_ensembl")
gene <- getBM(attributes=c('ensembl_gene_id','external_gene_name'),
filters = 'ensembl_gene_id',
values = results$ID,
mart = ensembl)You can use the merge function to bring the DEGRE output information
of the genes. Attention: you need to rename the columns of the gene data
frame in a way that the column with the gene IDs will have the same name
in both data frames: results from the DEGRE function (DEGRE
package) and gene from the getBM function (biomaRt
package).
colnames(gene)[1] <- "ID"
DEGREresults_gene_name <- merge(results, gene, by = "ID")The data frame DEGREresults_gene_name now has the gene
names from ensembl and all the information of the DEGRE function
output.
The DEGRE package has the VolcanoDEGRE function to visualize the proportion of downregulated and upregulated genes by applying a log2FC cutoff.
Here follows the quick start of the function with default parameters:
VolcanoDEGRE(results = results,
log2FC_cutoff = 1,
padj = 0.05,
delabel = "",
font.x = 10,
font.y = 10,
font.tickslab = 10,
downregulated_color = "coral2",
upregulated_color = "cornflowerblue",
xlab = "log2Foldchange",
ylab = "-log10(P-value)",
legend_position = "right",
legend.title = "Regulation")In the results argument, you must specify the data frame
you want to plot. The following result is shown below.

We recommend you to save it using the png function with the following parameters:
png(filename = "Volcano_DEGRE_example.png",
width = 1700,
height = 1200,
res = 350)
# Then, you paste the code to do the bar plot.
Paste here the code to do the bar plot and run it.
# And so, you close the plotting:
dev.off()Be free to change any of the arguments on the VolcanoDEGRE function.
The explained parameters are described below:
results - a data.frame object. It
receives the output of the DEGRE function, filtered or not, as
input.
log2FC_cutoff - it stores the cutoff of the
log2FoldChange.
padj - it stores the cutoff of the
P-adjusted value (Q-value).
font.x - the font size of the x axis.
font.y - the font size of the y axis.
font.tickslab - the font size of the ticks
lab.
downregulated_color - the colors of the
downregulated genes. The default is “coral2”.
upregulated_color - the colors of the upregulated
genes. The default is “cornflowerblue”.
xlab - the x lab text. The default is
“log2Foldchange”.
ylab - the y lab text. The default is
“-log10(P-value)”.
legend_position - you need to specify here the
position of the legend. The default is “right”.
legend.title - the title of the legend.
The DEGRE package has the BarGraphDEGRE function to get a bar plot showing the number of downregulated and upregulated genes.
Here follows the quick start of the function with default parameters:
BarGraphDEGRE(results = results,
log2FC_cutoff = 1,
downregulated_color = "coral2",
upregulated_color = "cornflowerblue",
xlab = "Regulation",
ylab = "Number of genes",
font.x = 10,
font.y = 10,
font.tickslab = 10,
legend_position = "right",
legend.title = "Regulation")At the results argument, you must specify the data frame
you want to plot. The following result is shown below.

We recommend you to save it using the png function with the following parameters:
png(filename = "BarPlot_DEGRE_example.png",
width = 1500,
height = 1200,
res = 350)
# Then, you paste the code to do the bar plot.
Paste here the code to do the bar plot and run it.
# And so, you close the plotting:
dev.off()Be free to change any of the arguments on the BarGraphDEGRE function.
The explained parameters are described below:
results - a data.frame object. It receives
the output of the DEGRE function, filtered or not, as input.log2FC_cutoff - it stores the cutoff of the
log2FoldChange.downregulated_color - the bar color related to the
number of downregulated genes. The default is “coral2”.upregulated_color - the bar color related to the number
of upregulated genes. The default is “cornflowerblue”.xlab - the x lab text. The default is
“Regulation”.ylab - the y lab text. The default is “Number of
genes”.font.x - the font size of the x axis.font.y - the font size of the y axis.font.tickslab - the font size of the ticks lab.legend_position - you need to specify here the position
of the legend. The default is “right”.legend.title - the title of the legend.