

A long journey is best broken into small steps, and the importance of taking a rest must never be underestimated.
The currr package is a wrapper for the
purrr::map() family but extends the iteration process with
a certain number of checkpoints (currr =
checkpoints + purrr), where the evaluated
results are saved, and we can always restart from there.
Implementations of the family of map() functions with a frequent
saving of the intermediate results. The contained functions let you
start the evaluation of the iterations where
you stopped (reading the already evaluated ones from the
cache), and work with the currently evaluated
iterations while the remaining ones are running in a background
job. Parallel computing is also easier with the
workers parameter.
install.packages("currr")The following example uses currr to present an everyday
issue: run a time-demanding iteration, but you want to rerun it
again.
library(tidyverse)
library(currr)
options(currr.folder = ".currr", currr.wait = Inf)
# folder in your wd, where to save cache data
avg_n <- function(.data, .col, x) {
# meaningless function that takes about 1 sec
Sys.sleep(1)
.data |>
dplyr::pull({{ .col }}) |>
(\(m) mean(m) * x) ()
}tictoc::tic(msg = "First evaluation")
cp_map(.x = 1:50, .f = avg_n, .data = iris,
.col = Sepal.Length,
name = "iris_mean") |>
head(3)
#> [[1]]
#> [1] 5.843333
#>
#> [[2]]
#> [1] 11.68667
#>
#> [[3]]
#> [1] 17.53
tictoc::toc() # ~ 1:50 => 50 x 1 sec
#> First evaluation: 50.351 sec elapsed
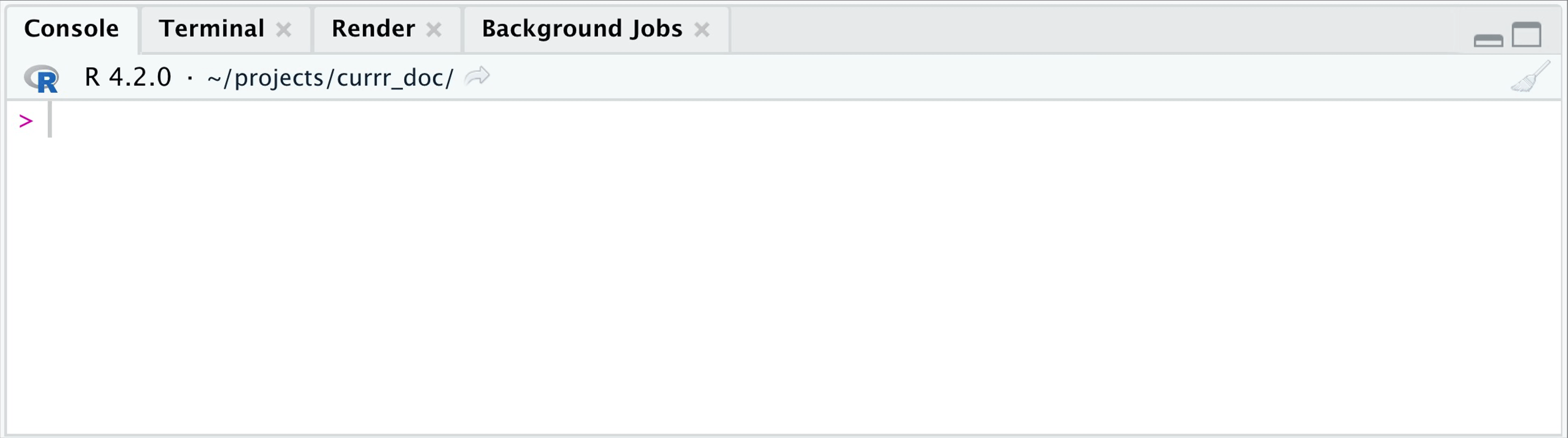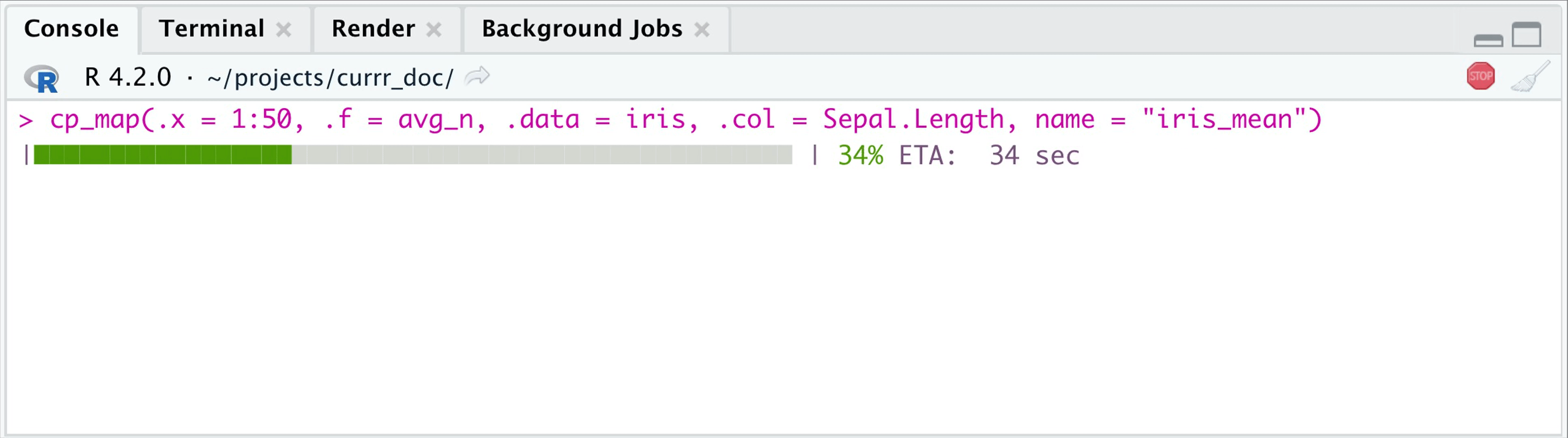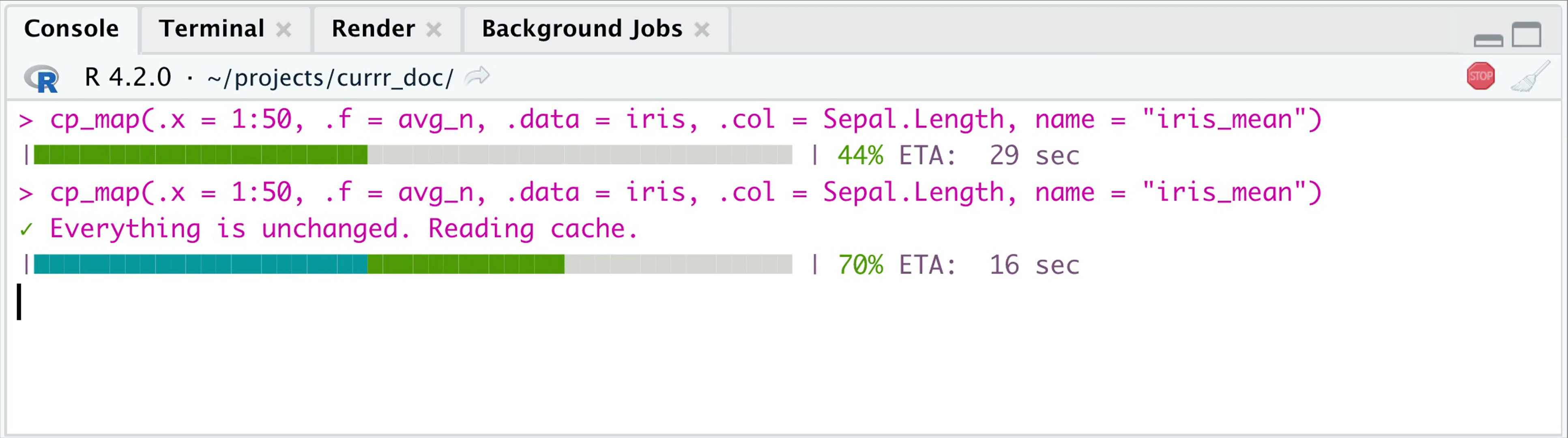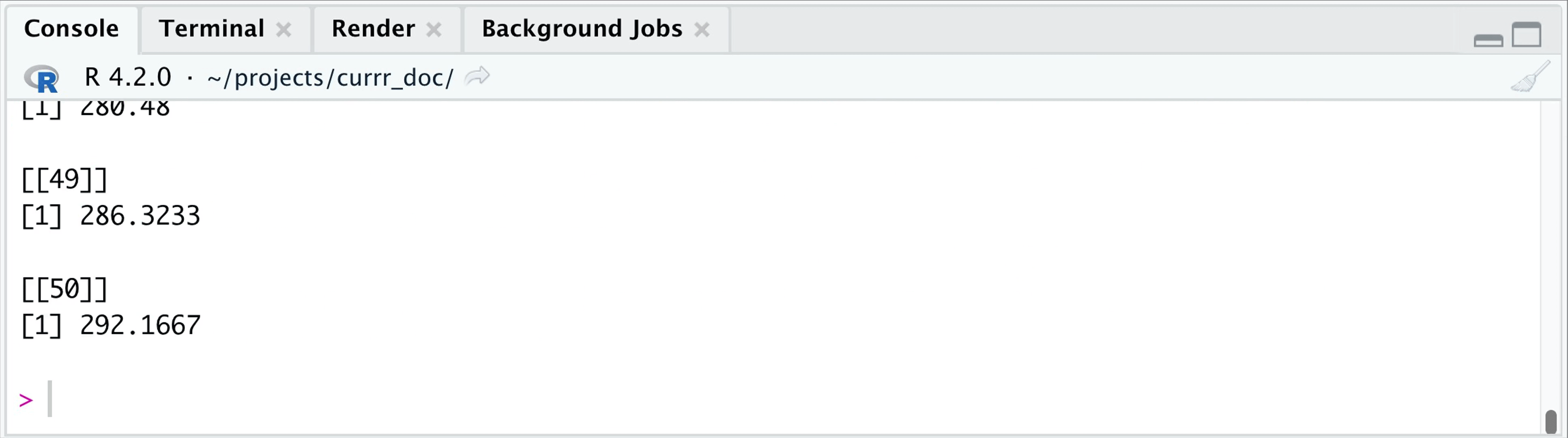
tictoc::tic(msg = "Second evaluation")
cp_map(.x = 1:50, .f = avg_n, .data = iris,
.col = Sepal.Length,
name = "iris_mean") |>
head(3)
#> ✓ Everything is unchanged. Reading cache.
#> [[1]]
#> [1] 5.843333
#>
#> [[2]]
#> [1] 11.68667
#>
#> [[3]]
#> [1] 17.53
tictoc::toc() # ~ 0 sec
#> Second evaluation: 0.034 sec elapsedIf the .x input and .f are the same, then
the 2nd time you call the function, it reads the outcome from the
specified folder (.currr). Also if .x changes,
but some of its part remain the same, then that part is taken from the
previously saved results, and only the new elements of .x
are called for evaluation. (If .f changes, then the process
will start from zero.)
tictoc::tic(msg = "Partly modification")
cp_map(.x = 20:60, .f = avg_n, .data = iris,
.col = Sepal.Length,
name = "iris_mean") |>
head(3)
#> ⚠ .x has changed. Looking for mathcing result to save them as cache
#> ◌ Cache updated based on the new .x values
#> [[1]]
#> [1] 116.8667
#>
#> [[2]]
#> [1] 122.71
#>
#> [[3]]
#> [1] 128.5533
tictoc::toc() # ~ 50:60 => 10 x 1 sec
#> Partly modification: 10.378 sec elapsedYou can remove the cache files, if you want to reset the process (or remove the already unnecessary files from your folder).
# only cache files for iris_mean
remove_currr_cache("iris_mean")
# all cache files
remove_currr_cache()You can also use multicore process (built on the
parallel package). After evaluation, the computation will
automatically reset to sequential.
options(currr.workers = 5) # <
tictoc::tic(msg = "Parallel computation")
cp_map(.x = 1:50, .f = avg_n, .data = iris,
.col = Sepal.Length,
name = "iris_mean") |>
head(3)
#> [[1]]
#> [1] 5.843333
#>
#> [[2]]
#> [1] 11.68667
#>
#> [[3]]
#> [1] 17.53
tictoc::toc() # ~ 50 / 5 => 10 sec
#> Parallel computation: 21.159 sec elapsedThis is another functionality that makes currr to be
cool. Working in RStudio you can set the wait parameter to
0-1/1+, define how many iterations you want to wait, and then let R work
on the remaining iterations in the background, while you can work with
the evaluated ones. If wait < 1, then it is interpreted as what
proportion of the iterations you want to wait. Whenever you recall the
function, it will return the already evaluated ones (use the
fill parameter to specify whether you want to get
NULLs to the pending ones.)
options(currr.wait = 20, currr.fill = FALSE)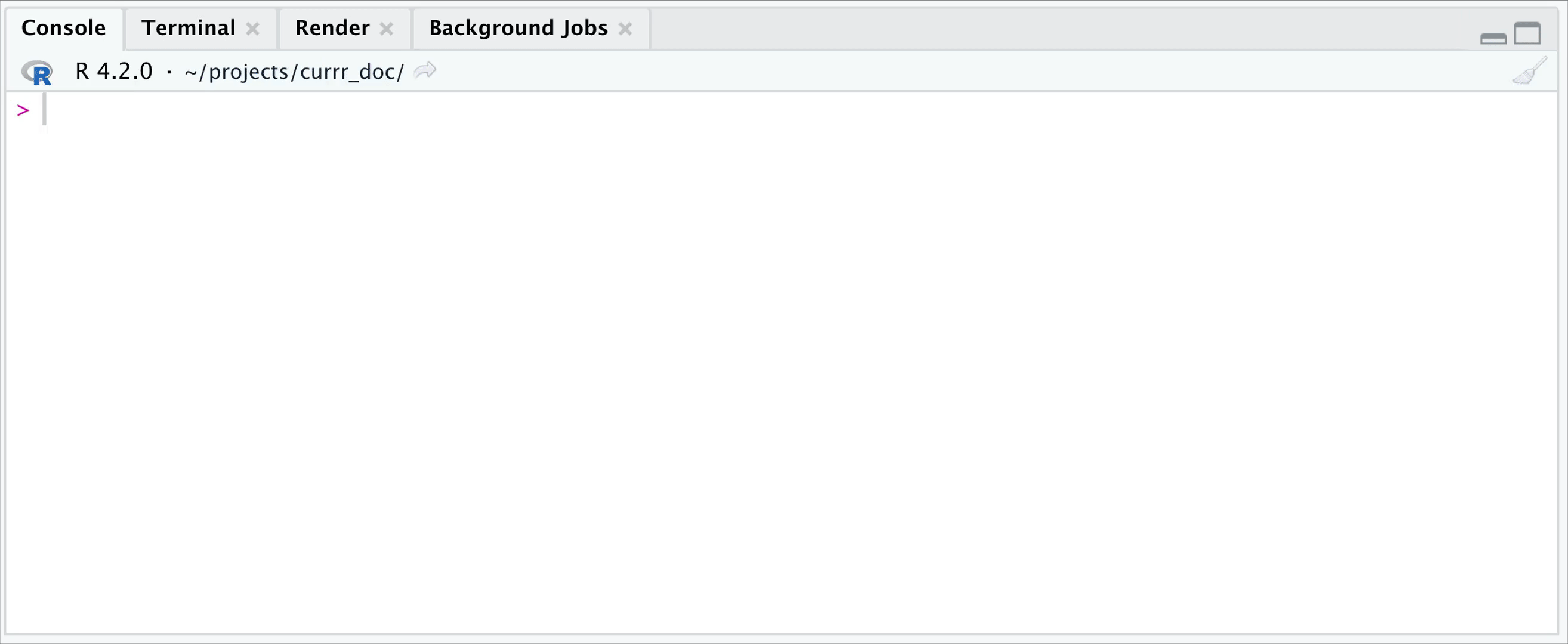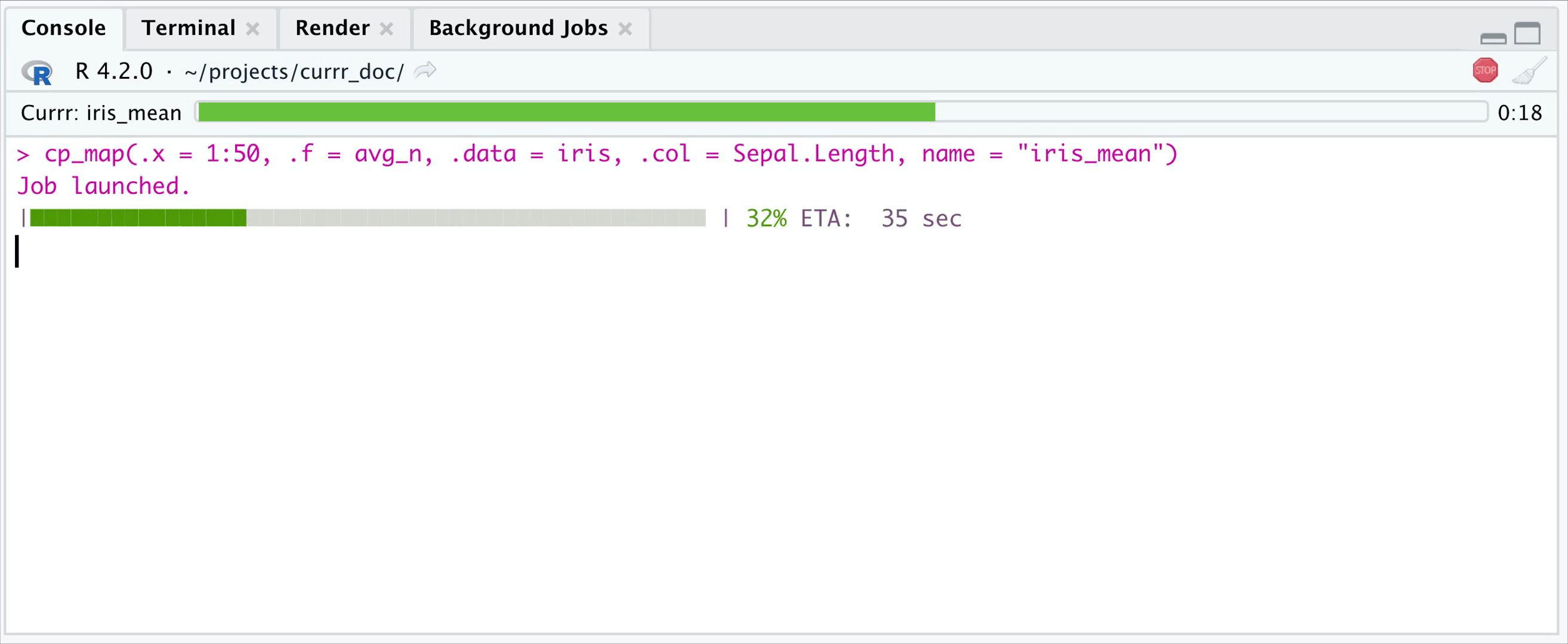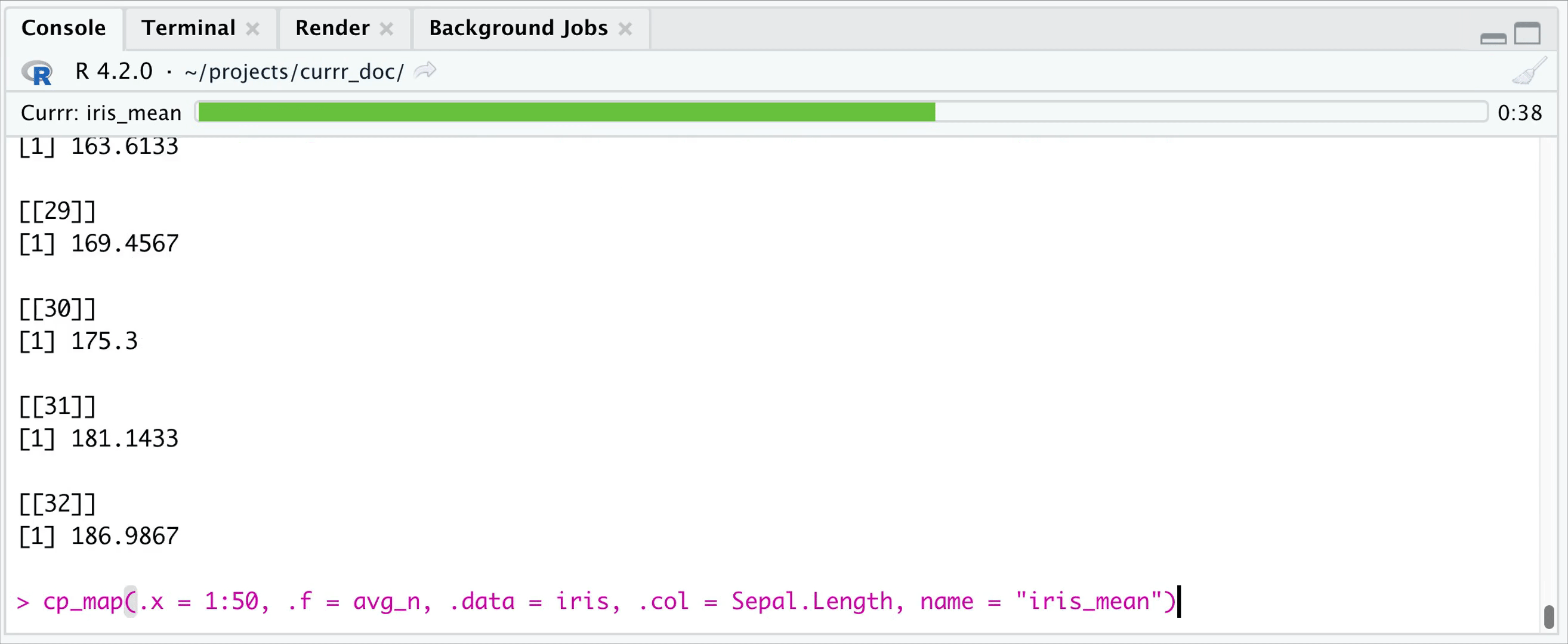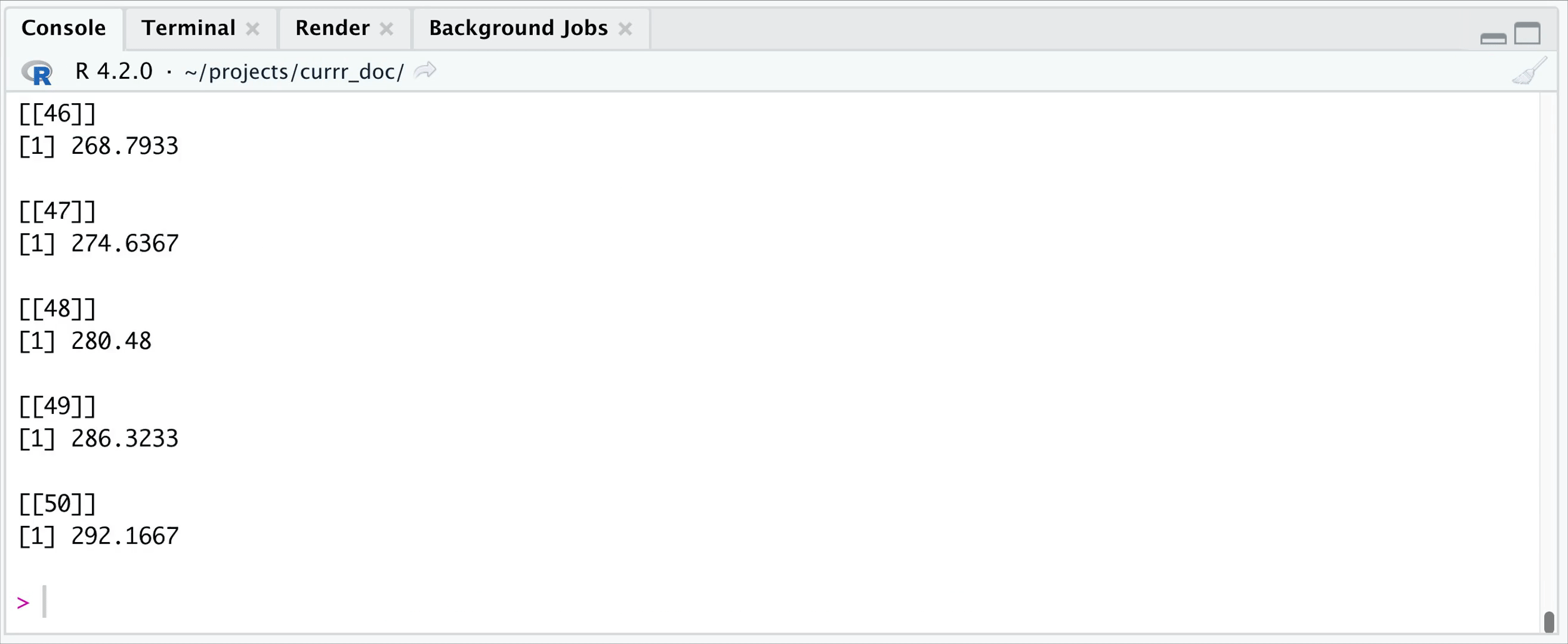
In the example above, you get your results, when 20 iterations are evaluated, but the job in the background keeps running.